Highly Parallel Enzymatic DNA Synthesis Using Electrowetting Droplet Technology
This technology is a method for synthesizing nucleic acids and other biopolymers without the use of a template. More specifically, this method is a highly parallel enzymatic synthesis of biopolymers on a microfluidic device. This technology has implications in DNA-based memory and RNA and DNA-based vaccine products.
Researchers
-
automated methods for scalable, parallelized enzymatic biopolymer synthesis and modification using microfluidic devices
United States of America | Published application -
automated methods for scalable, parallelized enzymatic biopolymer synthesis and modification using microfluidic devices
United States of America | Granted | 11,851,651
Figures
Technology
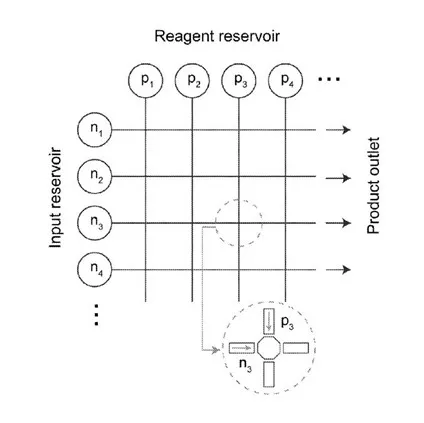
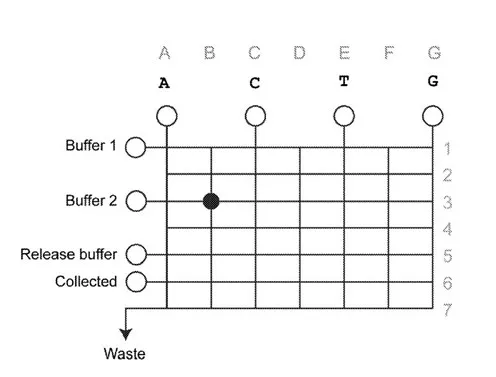
This technology is a method to synthesize nucleic acids in the absence of a template on a microfluidic chip. The microfluidic chip is comprised of fluid reservoirs containing: component building blocks (one for each of the nucleobases found in DNA) in suitable buffers such as physiological saline; the enzyme catalyst Tdt; and an initiation sequence (e.g., ssDNA sequence > 2 nucleotides) that contains a recognition site for TdT. To achieve nucleic acid synthesis on the microfluidic chip:
-
Droplets are simultaneously split from the reservoir containing TdT and one or more nucleobase reservoirs.
-
The droplets are then merged to form a combined droplet, which is incubated for at least 1 minute to allow TdT to attach nucleobases (a typical incubation time for catalysis of attachment of a nucleobase to the nucleic acid by TdT is 10 minutes at 37 C).
-
Next, the combined droplet is moved to a location containing the next nucleobase.
-
The previous steps are repeated until the desired nucleic acid has been synthesized. It can then be released by low-salt, heating, or cleaving with a nuclease-specific cut site incorporated 5’ to the initiation sequence.
The controlled movements of droplets on the microfluidic chip are carried out on a computer-readable medium such that the chip is rendered as a grid, and the intersection of grid lines are read as distinct locations. Further, this technology is not limited to synthesis of nucleic acids, but other biopolymers as well.
Problem Addressed
The two major approaches to DNA synthesis are chemical synthesis using phosphoramidite and enzymatic synthesis using Terminal deoxynucleotidyl Transferase (TdT). Chemical synthesis using phosphoramidite though a highly efficient technology is still relatively limited in the length of nucleotides they are able to synthesize (< 300 nucleotides in length). Enzymatic synthesis using TdT which is often used as an alternative to phosphoramidite, is a template-free method that works by affixing to an initiator and then initializing polymerization. However, this method is often less controlled compared to methods using phosphonamidite, which are able to add nucleotides in a more controlled step-wise manner.
By combining approaches of template-free DNA synthesis with current chemical approaches using microfluidic chips, this technology seeks to improve controllability of DNA synthesis while also allowing synthesis of larger oligonucleotides.
Advantages
-
Multiple ways to control movement of droplets – temperature and magnetic methods can be incorporated on the dielectric chip for synthesis.
-
Adaptable for implementation on any device that actuates microfluidic flow across a chip – examples include EWOD, acoustic droplet distribution, volumetric displacement distribution, ink-jet type fluidic distributors, etc.
-
Parallelized DNA synthesis – allows for each location on the chip to independently grow a long single-stranded DNA.
-
Programmable – movements of droplets containing the nucleotide can be directed to any location on the chip.
-
Synthesis of large oligonucleotides – oligonucleotides having up to 10,000 bases can be synthesized.
License this technology
Interested in this technology? Connect with our experienced licensing team to initiate the process.
Sign up for technology updates
Sign up now to receive the latest updates on cutting-edge technologies and innovations.