High-Throughput Native Context Mapping of the Regulatory Genome
This invention is a high-throughput CRISPR/Cas9-based approach that analyzes the regulatory genome for function in its native context. Understanding gene regulation for improved control allows for a number of applications including better management of stem cell differentiation.
Researchers
-
high-throughput crispr-based library screening
United States of America | Granted | 11,306,308
Figures
Technology
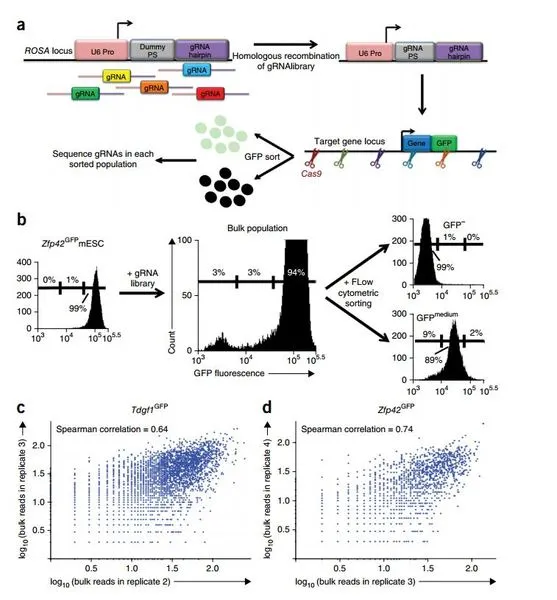
CRISPR/Cas (has been used in genome-wide mutation screens to identify genes required for survival, drug resistance, and tumor metastasis. MERA improves upon this previous gene mutation screening approach by ensuring cells receive a precise number of guide RNA (gRNA) per cell (i.e., one or more than one for combinatorial studies) and allows for gRNA libraries to be used without any laborious molecular cloning into a delivery vector. These methods can be used to screen for the effect of mutations anywhere in the genome.
Problem Addressed
Gene regulation, which underlies human variation, disease, and cancer, is poorly understood making it difficult to predict the effects of cis-regulatory variants on gene expression and to predictively alter gene expression during stem cell differentiation and reprogramming. Currently, there is no high-throughput approach capable of determining the relative importance of each gene regulatory element on native gene expression levels. This technology, Multiplexed Editing Regulatory Assay (MERA), enables the analysis of the regulatory genome at single base resolution in its native context.
Advantages
- Single base resolution
- No repeated and laborious cloning steps
- High-throughput
Publications
Rajagopal, N., et al. "High-Throughput Mapping of Regulatory DNA." Nature Biotechnology 34 (2016): 167–174. doi: 10.1038/nbt.3468.
License this technology
Interested in this technology? Connect with our experienced licensing team to initiate the process.
Sign up for technology updates
Sign up now to receive the latest updates on cutting-edge technologies and innovations.