Deaminase-based RNA Sensors
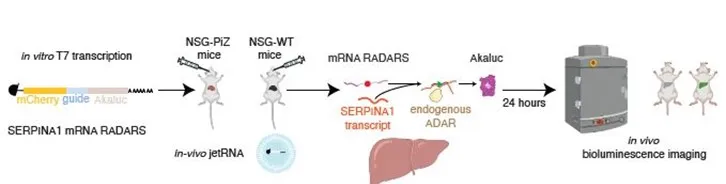
This technology is a novel RNA sensor system designed to express, detect and quantify specific RNA molecules in vivo without the need for genome engineering. It leverages the activity of adenosine deaminase acting on RNA (ADAR), along with a single-stranded RNA (ssRNA) sensor, to enable RNA-responsive expression of the payloads, such as reporters or therapeutic proteins. Traditional methods often rely on transgenic models or tagging strategies that alter native cellular processes, whereas this system enables cell-type-specific RNA sensing with minimal perturbation. The technology offers a powerful new platform for controlled expression of an exogenous RNA based on the presence of an endogenous transcript as well as means for studying gene expression and cell behavior in vivo within a more physiologically relevant context.
Researchers
-
deaminase-based rna sensors
Singapore | Pending -
deaminase-based rna sensors
Hong Kong | Published application -
deaminase-based rna sensors
United States of America | Granted | 12,195,734 -
deaminase-based rna sensors
United States of America | Published application -
deaminase-based rna sensors
European Patent Convention | Published application -
deaminase-based rna sensors
Japan | Published application -
deaminase-based rna sensors
Australia | Pending -
deaminase-based rna sensors
Canada | Published application -
deaminase-based rna sensors
China | Published application -
deaminase-based rna sensors
Korea (south) | Published application -
deaminase-based rna sensors
Israel | Published application -
deaminase-based rna sensors
Mexico | Pending -
deaminase-based rna sensors
New Zealand | Pending
Figures
Technology
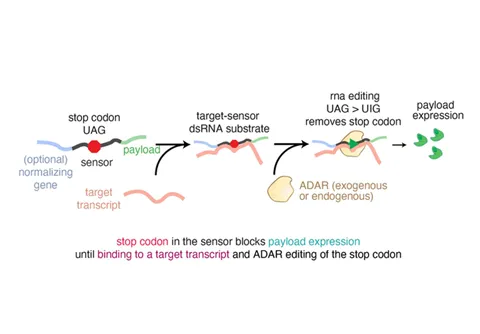
The system consists of an ssRNA sensor containing a stop codon and a payload sequence, along with an ADAR deaminase. When the sensor encounters its complementary ssRNA target, it hybridizes to form a double-stranded RNA (dsRNA) structure. A deliberate mismatch—typically an adenine:cytidine mispairing—at the stop codon site creates a substrate for ADAR. ADAR edits the mismatched adenosine to inosine, which is interpreted as guanosine during translation. This editing event converts the stop codon into a sense codon, allowing downstream translation of the payload. As a result, expression of the reporter or functional protein reflects the presence of the target RNA, enabling dynamic, quantitative RNA sensing in living cells.
Problem Addressed
This technology addresses the major limitations of current RNA tracking and manipulation techniques, which typically require genome manipulation, tagging, or the use of transgenic organisms. Such approaches can alter gene expression patterns and disrupt normal cellular activity, making it difficult to study RNA dynamics in a native context. Additionally, creating transgenic organisms is time-consuming, costly, and often not feasible in many model systems. This RNA sensor system enables precise, in vivo RNA measurement and cell-type-specific manipulation without the need for permanent genetic modifications or genomic integration.
Advantages
-
Capable of real-time, in vivo monitoring of RNA expression levels with potential reversibility and tunability.
-
Minimizes cellular perturbation by avoiding overexpression artifacts and preserving native gene regulation.
-
Allows for selective RNA sensing and payload expression in specific cell populations based on endogenous RNA expression patterns.
-
Compatible with a range of payloads, including fluorescent reporters (e.g., EGFP, luciferase), enzymes, transcription factors, and therapeutic proteins.
-
Can utilize endogenous or engineered ADAR variants, including Cas13-ADAR fusion proteins, for enhanced specificity and programmability.
Publications
U.S. Patent No. 12,195,734, Deaminase-based RNA sensors, Abudayyeh et. al, issued Jan. 14, 2025.
USSN 18/972,398, Deaminase-based RNA sensors, Jiang et. al, filed Dec 6, 2024
Jiang, K., J. Koob, X. D. Chen, R. N. Krajeski, Y. Zhang, V. Volf, W. Zhou, S. R. Sgrizzi, L. Villiger, J. S. Gootenberg, F. Chen, and O. O. Abudayyeh. "Programmable Eukaryotic Protein Synthesis with RNA Sensors by Harnessing ADAR." Nature Biotechnology 41, no. 5 (May 2023): 698–707. https://doi.org/10.1038/s41587-022-01534-5.
PCT/US2022/033459, Deaminase-based RNA sensors, Abudayyeh et. al, filed June 14, 2022.
License this technology
Interested in this technology? Connect with our experienced licensing team to initiate the process.
Sign up for technology updates
Sign up now to receive the latest updates on cutting-edge technologies and innovations.