Single Cell Super-Resolution Proteomics
Patch2MAP uses super-resolution microscopy to label and image subcellular structures of neurons and the associated protein distributions. This method characterizes cells by their electrical activity while preserving cellular integrity, allowing for novel investigations into the fine structure and protein content of brain cells. This method opens new avenues to investigate human health and disease by combining molecular, structural, and functional approaches
Researchers
-
single cell super-resolution proteomics
Patent Cooperation Treaty | Pending
Figures
Technology
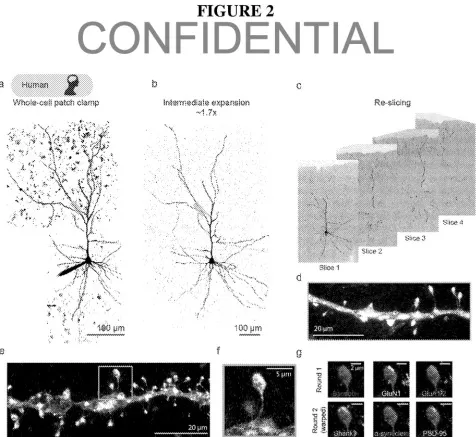
Patch2MAP analysis can be conducted on any brain tissue sample with viable neuronal tissue. The method involves conducting the patch-clamp procedure on one or more of the neurons to record their individual electrical activity. Next, the patch-clamped neurons are filled with a dye to identify and highlight morphological features. The eMAP protocol was modified to achieve super-resolution proteomic imaging of the filled cells. This alteration allows for the recovery of the dye’s intracellular distribution. The tissue is then treated with anywhere from 2-30 antibodies which target desired proteins and expanded to visualize the structure.
Problem Addressed
This technology allows for the full structural labeling of individual neurons for subsequent protein analysis. Current methods for analyzing the structure and function of sub-cellular processes use super-resolution microscopy. However, these approaches require inducing cells to express a foreign protein as a label for identification. This is typically done through genetic modification or viral transduction. In species or cells where, genetic medication is not feasible (most importantly the human brain), current state of the art techniques is unable to visualize proteins and assign them to the appropriate cell types and their subcellular compartments.
Patch2MAP overcomes these limitations by combining patch-clamp technology with epitope preserving magnified analysis of proteome (eMAP). Patch-clamp electrophysiology fills cells with an electrolyte solution to measure the flow of electrical currents across cell membranes. This can be used to study the biophysical properties of cells and identify cells by their electrical activity. eMAP is a synthetic gel-based super-resolution imaging technique which achieves ultrastructural imaging by coupling physical tissue expansion with common microscope techniques. Through the gelation process, proteins are anchored in place in a gel-based network. Tissue can then be expanded while the molecular structure remains in the same relative place. The novel Patch2MAP technology combines the benefits of both eMAP and patch-clamp electrophysiology, while eliminating the requirement of cells to express foreign proteins.
Advantages
- Capable of full morphological labeling of individual neurons from any species or cell type
- Allows for morphological reconstruction of fine processes including dendritic spines and axons
- Enables multiple rounds of de-staining and re-staining the tissue without loss of structural integrity for investigation of many proteins
Publications
Vardalaki, D., et al. "Patch2MAP Combines Patch-Clamp Electrophysiology with Super-Resolution Structural and Protein Imaging in Identified Single Neurons Without Genetic Modification." bioRxiv [Preprint], March 21, 2023. Accessed February 14, 2022. https://doi.org/10.1101/2023.03.20.533452.
License this technology
Interested in this technology? Connect with our experienced licensing team to initiate the process.
Sign up for technology updates
Sign up now to receive the latest updates on cutting-edge technologies and innovations.