Structured DNA Conjugates
This technology is a method of creating custom 3D polyhedral DNA nanostructures. These nanostructures can be designed to mimic biological structures such as virus capsids, toxins, protein assemblies, lipids, and sugars. Therefore, this technology has many potential applications including drug delivery, immuno-stimulation for vaccines, and biosensing for diagnostic use.
Researchers
-
stable nanoscale nucleic acid assemblies and methods thereof
European Patent Convention | Granted | 3,448,997 -
stable nanoscale nucleic acid assemblies and methods thereof
United States of America | Granted | 11,410,746 -
stable nanoscale nucleic acid assemblies and methods thereof
United Kingdom | Granted | 3,448,997 -
stable nanoscale nucleic acid assemblies and methods thereof
France | Granted | 3,448,997 -
stable nanoscale nucleic acid assemblies and methods thereof
Germany | Granted | 3,448,997 -
stable nanoscale nucleic acid assemblies and methods thereof
Switzerland | Granted | 3,448,997 -
stable nanoscale nucleic acid assemblies and methods thereof
Netherlands | Granted | 3,448,997 -
stable nanoscale nucleic acid assemblies and methods thereof
Ireland | Granted | 3,448,997 -
stable nanoscale nucleic acid assemblies and methods thereof
Belgium | Granted | 3,448,997 -
stable nanoscale nucleic acid assemblies and methods thereof
United States of America | Published application
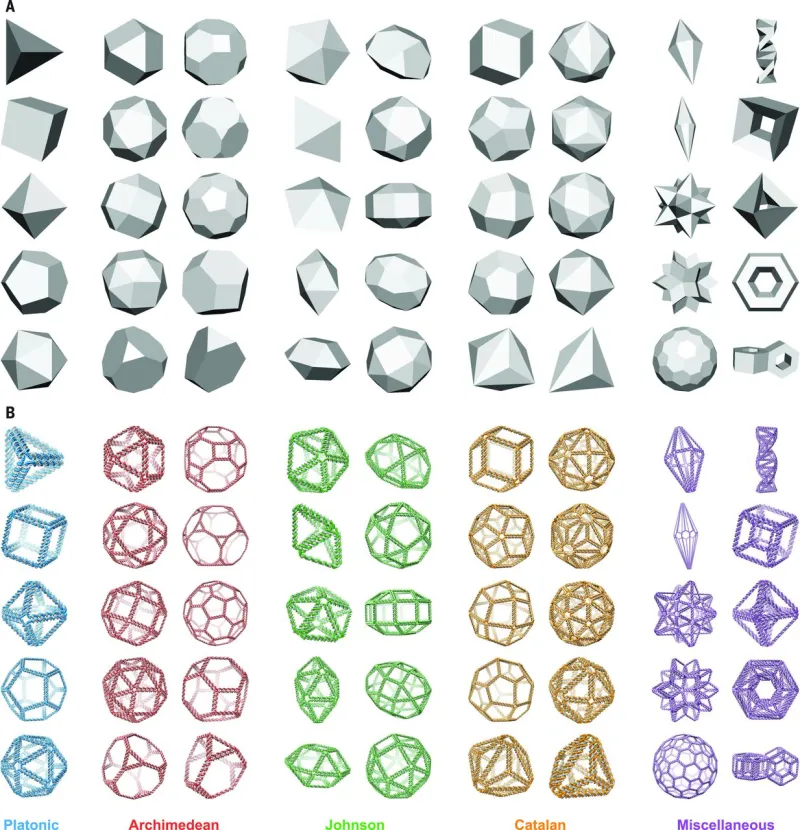
Figures
Technology
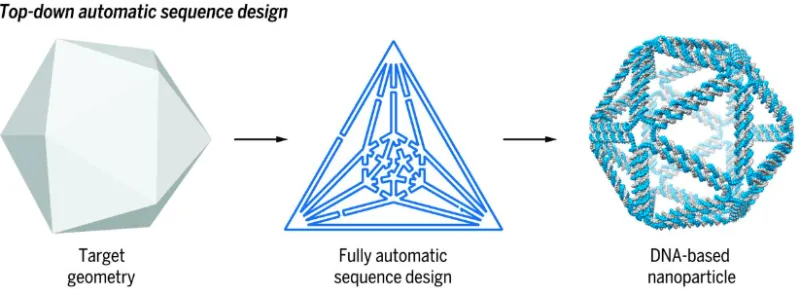
This technology describes a method for the top-down design of nucleic acid nanostructures based just on target shape. This approach involves first choosing a polyhedral form as the target structure and the desired geometric parameters and physical dimensions of it. Geometric parameters like vertex, face, and edge information can be determined from a polyhedral wire-mesh model of the target shape. The process continues by establishing the route of the single-stranded nucleic acid scaffold that traces throughout the structure, and finally determining the sequences of the scaffold strand and staple strands. Identifying the route of the scaffold is a multi-step process that makes use of a spanning tree, which establishes connectivity between all the vertices in the nanostructure. Once the spanning tree is generated, it can be used to produce a Eulerian circuit (a trail in a graph that visits every edge exactly once and starts and ends on the same vertex). Asymmetric polymerase chain reaction (PCR) is used to make the nanostructures from the template nucleic acid scaffold. The nanostructures can contain one or more moieties, such as proteins, lipids, small molecules, and RNAs bound to them for use in various applications.
The patent for this technology has been issued in Europe as 3448997 and validated in the United Kingdom, France, Germany, Switzerland, Netherlands, Ireland, and Belgium.
Problem Addressed
DNA nanostructures, also known as DNA origami, are 3D structures made of numerous short staple strands of DNA which guide the folding of a long scaffold polynucleotide strand into a custom polyhedral shape. There are currently several computational design tools that facilitate manual, bottom-up programming of DNA origami to form a desired shape using complementary Watson-Crick base pairing. However, only one approach tackles the inverse problem of using the geometry of a target shape to design a DNA nanostructure, and this method has its limitations. For example, it is only semi-automated and may produce unstable nanostructures that are unsuitable for many applications. Additionally, the geometries produced through this method must be spherical, restricting its scope. This MIT technology is fully automated and can produce a wide variety of non-spherical shapes, providing more versatility for applications with specific shape requirements.
Advantages
-
Fully automatic structure formation from only target shape
-
Generates many different topologies, not just spherical
-
Capable of modification pre- or post-assembly to be conjugated with other molecules
-
Wide range of potential applications provides flexibility to a licensing company
Publications
Rémi Veneziano, et al. "Designer Nanoscale DNA Assemblies Programmed from the Top Down." Science 352, 1534-1534 (2016). DOI: 10.1126/science.aaf4388.
License this technology
Interested in this technology? Connect with our experienced licensing team to initiate the process.
Sign up for technology updates
Sign up now to receive the latest updates on cutting-edge technologies and innovations.